This page is a part of the Computer Vision Wiki. The wiki is devoted to computer vision, especially low level computer vision, digital image analysis, and applications. The exposition is geared towards software developers, especially beginners. The wiki contains discussions, mathematics, algorithms, code snippets, source code, and compiled software. Everybody is welcome to contribute with this in mind - all links are no-follow.
Cell decomposition of images
From Computer Vision Wiki
A binary image is a combination of black and white pixels. Normally, it is assumed that objects are black and the background is white. How do you represent these objects? Well, each pixel is given by its location in the image, so it is just a pair of numbers. It seems natural to represent the object simply as a list of pairs of numbers and this is the way it is commonly done. We will follow this convention.
Beyond that however the question is how to think of pixels. The common view is that pixels are dots, let's call them pixel-dots. They are located on a grid. Of course to study the image one has to understand how these dots form objects. This requires the notion of adjacency among the pixel-dots. If two pixels are adjacent they are connected by an edge. The result is a graph representation of binary images.
This seems simple enough. However it turns out that the decision on which nearby pixel-dots are adjacent is not obvious. Should they be connected only along the grid? Or should the adjacency go diagonally too?
Both are equally legitimate answers.
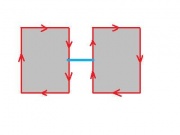
It may seem that choosing one of the two and sticking with it will provide a solid foundation. The trouble starts when one considers the complement of a set of pixels. The complement should also be a set of pixels with the same adjacency relations. But consider this.
If adjacency goes diagonally then A and D are connected. Then B and C are not connected, so adjacency does not go diagonally. Wait a minute...
This paradox can be resolved by having different adjacencies for the set and its complement. But, it's a can of worms... One has to create discrete geometry - from scratch. And things are still messy. Just imagine what happens in dimension 3! Read about this development elsewhere...
Our approach is very traditional, mathematically solid, and even philosophically satisfying!
The approach is "Euclidean" - the universe is a continuum.
Enough of philosophy.
Why? Because we want to break this continuous universe into small discrete pieces. Let's call them pixel-tiles. Since these pieces are discrete, we can study the universe with computers. On the other hand, put together pixel-tiles form a continuum. This continuum can be studied with the tools accumulated over the last 2 thousand years: Euclidean geometry, Newtonian physics and calculus, etc. For example, lines are lines not sequences of pixels.
Of course, the shapes of these tiles may be different. They may be squares, triangles, hexagons, etc. In fact they may have different shapes in the same image. The shapes of cells may even be curved, the approach remains valid. Generally, these tiles, and bricks, are called cells.
This gives us a uniform terminology for all dimensions:
- a vertex is a 0-cell,
- an edge is a 1-cell,
- a pixel is a 2-cell,
- a voxel is a 3-cell,
- etc.
Now the geometric picture is clear, we need to find a consistent way of treating the boundaries of pixels. This is important because pixels are attached to each other along their boundaries.
The way to address this issue is suggested by the list above. The boundary of a voxel consists of 6 faces, the boundary of the pixel consists of 4 edges, the boundary of the edge consists of 2 vertices. Therefore the decomposition of the image will contain all of these entities.
Note that we are not suggesting to every pixel is decomposed into a square, 4 edges, and 4 vertices. Edges and vertices are independent objects as they may be shared by adjacent pixels.
One advantage of this approach is that it is uniform throughout all dimensions:
- The boundary of an edge, 1-cell, consists of its two end-points, 0-cells;
- The boundary a pixel, 2-cell, consists of its four edges, 1-cells;
- The boundary of a voxel, 3-cell, consists of its six faces, 2-cells;
- etc.
Now, we are on the solid ground to address adjacency. As the image is made of pixels, they are attached to each other along the edges they share. Again, this it is easily applicable to all dimensions:
- two adjacent edges, 1-cells, share a vertex, 0-cell;
- two adjacent pixels, 2-cells, share an edge, 1-cell;
- two adjacent voxels, 3-cells, share a face, 2-cell;
- etc.
The hierarchy becomes clear.
Thus, our approach to image decomposition, in any dimension, boils down to the following.
A kind of cell decomposition commonly occurs in mesh generation. We are given a disorganized "point cloud". This is simply a collection of points, or vertices, or 0-cells, in space. Then some of the points are connected by edges, 1-cells. Finally, each triple of edges AB, BC, CA produces a triangle, or 2-cell.
The most dramatic advantage of this approach will become evident as we start doing algebra with cells. For that see Homology in 2D and Homology in 3D.
A more immediate consequence of (and a reason for) the decision to rely on cell decomposition is the simplification of the analysis algoritm. The algorithm is incremental. As pixels are added one by one, the topology of the image changes. Turns out, only adding edges needs attention. Since the new edge connects two vertices that have been added previously, there are just 3 cases to consider.
Now imagine that instead of an edge we are adding a whole pixel at once. It is adjacent to 8 other pixels. These are the consequences:
- There are 2^8 possible arrangements - one arrangement if you are adding an edge.
- These pixels may belong to up to 4 different objects - 2 objects if you are adding an edge.
- The number of cases to consider is hard to count - 3 cases if you are adding an edge.
Just imagine what happens in 3D!
There are other consequences such as the ease of computation of areas, perimeters etc.
Finally let's consider the way we record cells in images. Once again we stick with a familiar approach - via analytic geometry. Vertices are points so they can be recorded as pairs of integers. Edges are vectors, so an edge is recorded by its initial point and the direction - 4 integers. What about pixels? A pixel is recorded by the coordinate of its upper left corner - 2 integers. Observe that if the image is XxY, the coordinates of pixels run from 0 to X-1 (Y-1), while the coordinates of vertices run from 0 to X (Y).
Note: In the actual analysis algorithm this issue is treated slightly differently. First, an edge has length 1 and there are only 4 directions, so it is always (a,b,c,d) with (c,d) = (1,0), (-1,0), (0,1) or (0,-1). Second, vertices are represented as edges with no direction: (a,b,0,0).
Top finds
- Casinos Not On Gamstop
- Non Gamstop Casinos
- Casino Not On Gamstop
- Casino Not On Gamstop
- Non Gamstop Casinos UK
- Casino Sites Not On Gamstop
- Siti Non Aams
- Casino Online Non Aams
- Non Gamstop Casinos UK
- UK Casino Not On Gamstop
- Non Gamstop Casino UK
- UK Casinos Not On Gamstop
- UK Casino Not On Gamstop
- Non Gamstop Casino UK
- Non Gamstop Casinos
- Non Gamstop Casino Sites UK
- Best Non Gamstop Casinos
- Casino Sites Not On Gamstop
- Casino En Ligne Fiable
- UK Online Casinos Not On Gamstop
- Online Betting Sites UK
- Meilleur Site Casino En Ligne
- Migliori Casino Non Aams
- Best Non Gamstop Casino
- Crypto Casinos
- Casino En Ligne Belgique Liste
- Meilleur Site Casino En Ligne Belgique
- Bookmaker Non Aams
- カジノ ライブ
- онлайн казино с хорошей отдачей
- スマホ カジノ 稼ぐ
- ブック メーカー オッズ
- Top 3 Nhà Cái Uy Tín Nhất
- Trang Web Cá độ Bóng đá Của Việt Nam
- Casino En Ligne Avis
- Casino En Ligne France
- Casino En Ligne
- 꽁머니 토토
- Casino Online Non Aams
- Meilleur Casino En Ligne